RESEARCH ARTICLE
Doi: 10.5578/tt.6474
Tuberk Toraks 2015;63(4):235-242

Mdr ve Xdr Mycobacterium tuberculosis ?rneklerinden streptomisin direncinden sorumlu
rpsL ve rrs gen mutasyonlarının analizi
Mohammad ARJOMANDZADEGAN1, Somayeh GRAVAND2
1 Arak Tıp Bilimleri ?niversitesi, T?berk?loz ve ?ocuk İnfeksiyon Hastalıkları Araştırma Merkezi, Mikrobiyoloji B?l?m?,
Arak, İran
1 Department of Microbiology, Tuberculosis and Pediatric Diseases Research Center, Arak University of Medical Sciences,
Arak, Iran
2 Arak İslam ?niversitesi, Bilim ve Araştırma Fak?ltesi, Mikrobiyoloji B?l?m?, Arak, İran
2 Department of Microbiology, Islamic Azad University, Arak Science and Research Branch, Arak, Iran
?ZET
Mdr ve Xdr Mycobacterium tuberculosis ?rneklerinden streptomisin direncinden sorumlu rpsL ve rrs gen mutasyonlarının analizi
Giriş: Streptomisin, ?ok ilaca diren?li t?berk?loz tedavisinde en etkili ila?lardan birisidir. Bakterisidal aminoglikozid bir antibiyotiktir ve diren? giderek artmaktadır. Streptomisinin etki mekanizması aminoa?il tRNA'nın uzama fazında "A"ya bağlanmasını engeller?? ve sonu? olarak bakteriyel protein sentezini durdurur. Bu ?alışmada Mycobacterium tuberculosis'in klinik ?rneklerinde streptomisin direncinin hızlı incelemesi ?alışılmıştır.
Materyal ve Metod: Bu ?alışmada balgam ve k?lt?r pozitif 105 M. tuberculosis'ten 45 tane streptomisine diren?li ve duyarlı ?rnek se?ilmiş ve rrs ve rpsL genlerindeki mutasyonlar a?ısından ?alışılmıştır. Polimeraz zincir reaksiyonu (PCR) i?in kullanılan spesifik primerler rpsL 1, rpsL 2, rrsR ve Frrs olarak adlandırılmıştır. PCR ?r?nleri sekanslanmıştır.
Bulgular: rpsL geni i?in 504 bp bandı ve rrs geni i?in 1027 bp bandı g?steren PCR ?r?nleri, uygun primerlerin se?imi ve uygun amplifikasyon programını sağlamıştır. Streptomisine diren?li 26 ?rnekten 26'sında spsL geninde mutasyon ve birinde rrs geninde değişklik vardı. Bu ?alışmada streptomisine duyarlı 19 ?rnekte mutasyon saptanmadı.
Sonu?: Streptomisin direncinden spsL geninde 43 ve 88 kodonlarda ve nadiren rrs genindeki mutasyonlar sorumludur.?
Anahtar kelimeler: Mycobacterium tuberculosis, streptomisin, ila? direnci, rpsL geni, rrs geni
SUMMARY
Analysis of rpsL and rrs genes mutations related to streptomycin resistance in Mdr and Xdr clinical isolates of Mycobacterium tuberculosis
Introduction: Streptomycin is a bactericidal and aminoglycoside antibiotic. It is one of the most effective drugs for treatment of multi-drug Tuberculosis disease. Incidence of resistance is increasingly reported. Its action mechanism is by inhibition of binding aminoacyl tRNA to position "A" in elongation phase, which finally it causes to stop bacterial protein synthesis. In this study, resistance rapid investigation to streptomycin was conducted in clinical strains of Mycobacterium tuberculosis.
Material and Method: In this study, among 105 strains of phlegm-positive and culture-positive Mycobacterium tuberculosis, 45 strains of resistant and sensitive to streptomycin were selected for possible mutations examination in genes rrs and rpsL. Specific primers that used for PCR were named rpsL 1, rpsL 2 and rrsR, Frrs. PCR products were sequenced.
Results: PCR Products represents 504 bp band for gene rpsL and 1027 bp for gene rrs that shows proper selection of primers and determining an amplification appropriate program. From 26 resistant strains to streptomycin 26 strain have mutation in rpsL gene and 1 strain have alteration in rrs gene. In this study 19 strains were sensitive to streptomycin that have no mutation in these gene.
Conclusions: Streptomycin resistance is mainly related to mutation at codons 43 and 88 "rpsL" gene and to a lesser extent "rrs" that are the greatest cause of drug resistance to streptomycin.
Key words: Mycobacterium tuberculosis, Streptomycin, drug resistance, rpsL gene, rrs gene
INTRODUCTION
Tuberculosis was associated with mankind throughout history. With formation of societies and crowding of increased population, tuberculosis was marked as a cause for one fourth of mortality in 17th and 18th centuries in Europe, so that in industrialization in Europe, it was called white plague. Despite extending and developing technology science, especially medical science as well as relatively improvement of living standards, it remains as one of the most important and common factors of people mortality (1). It is an infectious disease with chronic nature and highly contagious that in most cases resulted from Mycobacterium tuberculosis. Frequent site of its involvement is lung, but it infects other members in one third of cases (2). This disease is a health problem for human societies, especially in developing societies. Within these countries, between 0.1% to 0.3% population are infected this disease each year, although disease prevalence was going to fall in early 20th century, but since 80s, disease outbreak were raised. It is very dangerous for human being so that World Health Organization (WHO) has been suggested this problem as a "global EMS" in 1991 (3).
Drug resistance to tuberculosis bacillus occurs mainly when there is a mutation in bacillus and the reason of resistance is incomplete or incorrect treatment that allows resistant bacilli mutant turn into dominated strains. The incidence of multi-drug resistant tuberculosis (MDR-TB) is rapidly increasing and its incidence estimated at 460.000 worldwide in 2005 (4). If a person who is infected with TB is not treated, on average, he/she infects 10 to 15 people each year, because TB transmission is through respiratory tract. Drug-resistant incidence caused by increasing anti-tuberculosis drugs consumption and other factors are among major concerns. Streptomycin is from aminoglycosides family. Aminoglycosides inhibit protein synthesis by binding to ribosome, thus they have a bactericid effect (5). Streptomycin operation mechanism is that this drug acts on 16S rRNA and S12 ribosomal protein and this leads to ribosomal changes induction. These changes cause to make a mistake in reading mRNA and finally inhibition of protein synthesis (3,6,7)
Streptomycin resistance is mainly related to mutation in gene rpsL and in lesser amount to gene rrs, other factors such as changes in permeability of the wall and membrane and modifying enzymes of aminoglycosides are slightly effective. Mutation in gene rpsL depends on changes in codons 43 in 70% cases and codon 88 in much less cases. Streptomycin resistance occurs because of occurrence of mutation in genes such as rpsL and rrs that are respectively coding of 16SrRNA and ribosomal protein S12. They are responsible for high levels of resistance to Streptomycin (6,8,9,10).
MAterialS and METHODS
DNA Extraction?
CHELEX100 kit (Sigma) is used to DNA extraction. According its manufacture, 0/07 gr of CHELEX powder is saluted in 270 μl? TAE1X buffer and after adding a few new colonies of bacteria and severe vertex are incubated at 95?C for 45 minutes. CHELEX particles were fully separated during 3 stages. After heating and vortex at 14.000 rpm centrifuge for 10 minutes and final supernatant as DNA is used in PCR.
Primers Design
The used primers in the present study designed by OLIGO6, MEGA4, IDT software. Sequences the designed primer and PCR conditions to amplify target genes is presented in Table 1.
PCR
For rpsL gene PCR mixture containing 2 mL DNA (40 ng) 2.5 mL MgCl2, 2.6 mL rpsL F, rpsL R 0.7 macro liter of fourfold nucleotides desoxy and 0.7 mL of enzyme taq polymerase were used in a final volume of 25 mL.
For rrs gene, as above mentioned its desired primers was designed and PCR mixture containing 2 mL DNA (40 ng), 2.5 mL MgCl2, 2 mL rpsL F, rpsL R, 0.7 macro liter of fourfold nucleotides desoxy mixture and 1 mL of enzyme taq polymerase were used in a final volume of 25 mL.
PCR products were loaded on 1% agarose gel (Cinagen) and suitable molecular marker 100 bp (Vivantis) were used for band measurement.
Sequencing
PCR Products were sent to Source BioScience Company in order to DNA purification and sequencing process using Applied Biosysctem device. The results of sequencing were analyzed by Mega 4, Chromas and Bioedite software.
Mutation occurrence determination results were compared to resistance phenotypic results.
Results
From a total of 45 studied Mycobacterium tuberculosis strains, 26 srrain were streptomycin resistant and 19 of them were sensitive to drug. Running PCR on clinical samples, 504 bp band for rpsL? gene and 1027 band for rrs gene were provided in electrophoresis gel 1% that show correct choice of primers and determining an appropriate program for amplification (Figure 1 and 2).
Sequencing result showed that from 45 samples that were sent for sequencing of rpsL gene, all of 26 resistant samples had mutation in rpsL? gene and 19 sensitive samples had no mutation. Change in amino acid 121 included the highest amount of mutation. Change in position of Wobble site (third nucleotide) of codon occurred (AAA → AAG) i.e. (K-121-K).? Therefore, a lysine (K) amino acid turned into another of the same amino acid, so there is no mutation in protein surface and it is only on nucleotide level.
Some changes were recorded in codon 43 that is related to nucleotide 128 and lead to the conversion of AAG to AGG, that is, a change occurred in second nucleotide that lead to conversion of amino acid lysine to amino acid arginine (R). Thus for this change, there is a mutation in protein surface.
Some changes were recorded in codon 88 that is little but it is related to nucleotide 263 and lead to the conversion of AAG to AGG and this change occur in second nucleotide that leads to conversion of amino acid lysine to amino acid arginine. Thus for this change, there is a mutation in protein surface. In samples of 725.571 (AAG → A GG) and actually the K88R i.e. lysine is transformed into arginine. It means there are a mutation in codon 88 and a mutation in codon 43 (K43R), so these samples have mutations both in 43 of 88.
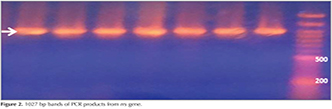
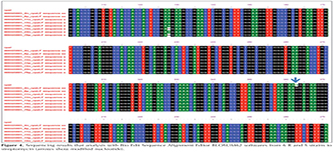
In Figure 3, a result of sequencing performed on a sample that is resistant to streptomycin is shown with Choromas and Mega-4 software. Results analysis of sequencing on resistant bacteria is shown that is done by using primers R sequence rpsL with Bioedit software in Figure 4.
18 resistant strains were sequenced for rrs gene. Among 18 samples, just a resistant sample (No. 84p) has mutation in rrs gene. 14 sensitive strains were reported without mutation.
Discussion
Tuberculosis is one of global problems and 3 million people lost their life due to this disease (11). In this study, mutations existed in rpsL and rrs genes in drug sensitive and resistant clinical strains were determined and compared with each other. Mutation in codons 43 (AAG → AGG) - 88(AAA → AAG) were reported as the most frequent mutations of rpsL gene worldwide. In present study, the most frequent mutated codons are codons 121 and 43 and 88.
Srinand sreevatsa was conducted a research to indentify mutations in rpsL and rrs in resistant mycobacterium to streptomycin, in this study, Tuberculosis mycobacterium 139 strains of patients in South America, Europe, Africa and Asia were examined that used sequence method and it was observed that 78 strains were resistant and 61 strains were susceptible that 54% of resistant strains had missense mutation in codon 43 of rpsL (k-43-R) also in codon 88, 4 Asian samples were detected. 4 strains from 8 strains that were got from Japan had mutations in (k-43-R) and had 2 strains in (k-88-Q), in addition just one strain isolated in New York among 78 resistant strains with mutations in rpsL? and rrs were together as mutations in rpsL was 56.8% and in rrs was 15.6% (6,12).
In a study that was conducted by Miho Fukuda in 1999, sequencing method was used. From 121 strains isolated, 26 strains were resistant to streptomycin, 78% mutation in codon 43 and 22% in codon 88 was from rpsL gene and no change was reported in rrs gene (7). In a study that was conducted by Ryan in 2004, sequencing method was used and it was shown that two rpsL and rrs genes are responsible for tuberculosis Mycobacterium resistance to streptomycin, results analysis showed that in 61% cases, rpsL? gene and in 24% cases, rrs gene are responsible for resistance. Using former reports that are conducted as an alternative in rrs and rpsL? genes, their results show that (52-57%) in rpsL? and (8-15%) in rrs had mutations and most highest mutation codon is codon 43 (Lys-Arg) (13).
In a study that was carried by WU Xue-Qiong in 2004, method of sequencing was used that 98 strains were studied, results from sequencing included 78 (79.6%) had mutation in codon 43 or 88 of rpsL gene (Lys> Arg), (6.1%) 6 in rrs gene were in (A-C 513) or (C-T 516) and (14.3%) 14 had no mutations (14).
Ozturk was conducted a research in 2005 as resistant molecular analysis to isoniazid, rifampin, and streptomycin and he used sequence method in which from total 52 samples that function of 5 were related to streptomycin and all had mutations in codon 43, therefore mutation in rpsL and rrs are responsible for over 70% of resistance to streptomycin which this result is consistent with present study (9,15).
In 2009, Shubhada indentified rapid resistance to isoniazid, rifampin and streptomycin among clinical isolates of Mycobacterium with mutation. Among them, 75% had mutations in rpsL? gene in codon 43. PCR multiplexing and sequence methods were referred in this study that mutations are seen in codons (43,88) rpsL gene and (491.513.516) in Mycobacterium tuberculosis. The largest mutation was in rpsL Lys43Arg that is 75% and in rrs gene was 12%, 2% had no mutation and 6% had mutation in position 516. 98% had phenotype DST compliance.
In his study, Pablo Bifani reviewed 26 strains using gene sequences, that 14 strains of them showed an amount of high level resistance to streptomycin and 6 sample were poly resistant, an analyze was performed to confirm that all isolated strains have a mutation in codon 43 of rpsL gene and change includes the conversion of lysine to threonine and no change was observed in rrs gene (16).
Tatjana Tracevska was conducted a research in 2004 that 66 strains were from TB patients that were resistant to Rif, NH, SM and 33 were resistant to EMB. (61%) 40 strains from 66 Isolated strain were resistant to streptomycin. From 26 remaining strains, (24%) 16 had a mutation at position 513 (A → C), 516 (C → T) of rrs gene and remained (15%) 10 of them were wild (17).
In a study in 2009 by Chaoui and colleagues using sequences method, from 58 strains isolated resistant to streptomycin, it was confirmed that most amount of mutations were in codon 43 of rpsL? gene and there was rare mutation in codon 512 of rrs gene. In present study also the most significant mutations were in codon 43 of rpsL gene (18).
In 2012, Arch Orthop Trauma Surg was done a comparison study between cultures L_J and absolute concentration with DNA sequencing for detection of drug resistant tuberculosis SPINAL/that only 7 drug resistant samples from 50 samples were recognized by culture and 34 days lasted, but by sequencing of 11 drug resistance, it was found that 6 days lasted. Majority of mutations in rpsL? genes were in codons 43 and 88 according to previous reports (19).
In 2012, in a research, Silke Feuerriegel conducted 97 strains to identify mutations in rpsL and rrs genes and so on by sequencing method, it results showed that none of strains resistant to streptomycin in rrs gene had mutation but 47.5% had resistant in rpsL? gene (20).
90-95% of mutations in mutants resistant to streptomycin had mutation in rpsL? and rrs that most of them happened in bp 43 88 bp. The largest mutation in rpsL in codon 43 is associated with a stop in protein synthesis in bacteria. Its consequences are fully consistent with DNA sequencing. There was no mutation in susceptible isolates and 84% of isolates showed resistant to streptomycin of mutation in rrs and rpsL genes. In this study, similar to other studies rpsL played a more important role than rrs in resistant to streptomycin, mutation in rspl occurs frequently in codon 43 and less in codon 88. 0.14% of isolates resistant in rrs, rpsL genes had no mutation. Findings in this study is consistent with results obtained in present study (3,15,21).
We sequenced 45 samples that were given to resistant streptomycin drug after PCR and electrophoresis on agarose gel. From 45 samples that were sent for sequencing of rpsL? gene, all of 26 resistant samples had mutation in rpsL? gene and 19 sensitive samples had no mutation. Also 18 resistant strains were sequenced for rrs gene. Among 18 samples, just a resistant sample has mutation in? rrs gene. Number of 14 sensitive strains were reported without mutation.
Among 45 of samples resistant to streptomycin that were sequenced, 17 samples had no any mutation in bp 43 88 bp region of rpsL gene. Also, 26 samples had mutations in this area that are listed in the table.
Mutations at codon AAA → AAG121, AAG → AGG43, 511 and 516 were reported as the most frequent mutations of gene l around the world. In present study, the most frequent mutated codons are codons 121 (40%) and codon 43 (20%) that in codon 121 as no change happened in slip position i.e. third nucleotide so amino acid does not change and lysine turn into lysine (AAA → AAG) thus no mutation happen in protein surface and there is only mutation in nucleotides surface. And actually, one codon of amino acid lysine is converted to another codon of the same amino acid.
Frequency of codon 43 is compatible with these results but frequency of codon 121 is inconsistent with these reports. Probably this is due to geographical differences between under study samples and this shows plenty of this mutation in samples. From 8 observed mutations in codon 43, all mutations are made under the effect of (AGG → AAG Arg) → AGG change and in codon 88 also a mutation as result of (Lis → AAG Arg) → AGG change and in codon 121 because of (Lys → AAA >> AAG) Lys) change.
But in 725.571 samples (AAG >> AGG) and actually K88R i.e. lysine is transformed to arginine. It means that there is a mutation in codon 88 and one in codon 43 (K43R) so theses samples have mutations in both 43 and 88. And they are valuable to us for assessment mutations. Sequencing method as a genotypic gold standard of molecular techniques were used. In this study, sequencing method was used to determine mutation in rpsL? gene as well as rrs gene.
REFERENCES
- Meya DB, McAdam KP. The TB pandemic: an old problem seeking new solutions J Intern Med 2007;261:309-29.
- Fair E, Hopewell PC, Pai M. International Standards for Tuberculosis Care: revisiting the cornerstones of tuberculosis care and control. Expert Rev Anti Infect Ther 2007;5:61-5.
- Finken M, Kirschner P, Meier A, Wrede A, B?ttger EC. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol Mol Microbiol 1993;9:1239-46.
- Zignol M, Wright A, Jaramillo E, Nunn P, Raviglione MC. Patients with previously treated tuberculosis no longer neglected. Clin Infect Dis 2007;44:61.
- Tracevska T, Jansone I, Nodieva A, Marga O, Skenders G, Baumanis V. Characterisation of rpsL, rrs and embB mutations associated with streptomycin and ethambutol resistance in Mycobacterium tuberculosis. Res Microbiol 2004;155:830-4.
- Sreevatsan S, Pan X, Stockbauer KE, Williams DL, Kreiswirth BN, Musser JM. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities Antimicrob Agents Chemother 1996;40:1024-6.
- Fukuda M, Koga H, Ohno H, Yang B, Hirakata Y, Maesaki S, et al. Relationship between genetic alteration of the rpsL gene and streptomycin susceptibility of Mycobacterium tuberculosis in Japan. J Antimicrob Chemother 1999;43: 281-4.
- Sreevatsan S, Pan X, Stockbauer KE, Williams DL, Kreiswirth BN, Musser JM. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob Agents Chemother 1996;40:1024-6.
- Ozturk CE, Sanic A, Kaya D, Ceyhan I. Molecular analysis of isoniazid, rifampin and streptomycin resistance in Mycobacterium tuberculosis isolates from patients with tuberculosis in D?zce, Turkey. Jpn J Infect Dis 2005;58:309-12.
- Morris S, Bai GH, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis 1995;171:954-60.
- Kam KM, Yip CW, Tse LW, Leung KL, Wong KL, Ko WM, et al. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol Lett 2006;256:258-65.
- Dobner P, Bretzel G, R?sch-Gerdes S, Feldmann K, Rifai M, L?scher T, et al. Geographic variation of the predictive values of genomic mutations associated with streptomycin resistance in Mycobacterium tuberculosis. Mol Cell Probes 1997;11:123-6.
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393:537-44.
- Wu XQ, Lu Y, Zhang JX, Liang JQ, Zhang GY, Li HM, et al Detection of streptomycin resistance in Mycobacterium tuberculosis clinical isolates using four molecular methods in China.Yi Chuan Xue Bao 2006;33:655-63.
- Via LE, Cho SN, Hwang S, Bang H, Park SK, Kang HS, et al. Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean Patients with drug-resistant tuberculosis. J Clin Microbiol 2010;48:402-11.
- Bifani P, Mathema B, Campo M, Moghazeh S, Nivin B, Shashkina E, et al. Molecular identification of streptomycin monoresistant Mycobacterium tuberculosis related to multidrug-resistant W strain. Emerg Infect Dis 2001;7:842-8.
- Si J, Wang Z, Wang Z, Li H. Sequencing-based detection of drug-resistant Mycobacteriu m tuberculosis in patients with spinal tuberculosis Arch Orthop Trauma Surg 2012 Jul;132:941-5.
- Chaoui I, Sabouni R, Kourout M, Jordaan AM, Lahlou O, Elouad R, et al. Analysis of isoniazid, streptomycin and ethambutol resistance in Mycobacterium tuberculosis isolates from Morocco. J Infect Dev Ctries 2009;3:278-84.
- Tracevska T, Jansone I, Nodieva A, Marga O, Skenders G, Baumanis V. Characterisation of rpsL, rrs and embB mutations associated with streptomycin and ethambutol resistance in Mycobacterium tuberculosis. Res Microbiol 2004 Dec;155:830-4.
- Feuerriegel S, Oberhauser B, George AG, Dafae F, Richter E, R?sch-Gerdes S, et al. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol 2012;12:90.
- Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol 2007;63:1096-106.
Yazışma Adresi (Address for Correspondence
Dr. Mohammad Arjomandzadegan
Department of Microbiology, Tuberculosis and
Pediatric Diseases Research Center,
Arak University of Medical Sciences,
ARAK - IRAN
e-mail: mmatinam81@yahoo.com